Epigenlab LLC was founded for development of new generation epigenetic DNA PCR tests for the early-stage detection of cancer based on new methods of DNA methylation assays by SibEnzyme.
In the tests we analyze DNA isolated from the vein blood and use a standard real-time PCR machine.
What is special in our approach?
We determine DNA methylation markers, but our DNA methylation assay is absolutely new.
Assay is based on a new type of enzymes, recently developed by SibEnzyme – methyl-directed DNA endonucleases or MD DNA endonucleases. MD endonucleases work like traditional restriction enzymes, but, on the contrary, they cut only methylated DNA and may be used in epigenetic studies. More details
GLAD-PCR assay – is a simple and inexpensive method for epigenetic studies of DNA preparations from clinical samples. It allows to detect even several DNA molecules with R(5mC)GY site of interest in a presence of excessive amount of unmethylated DNA. Just 3 easy steps in one tube. More details
GlaI-qPCR assay – is a simple and inexpensive method for epigenetic studies of DNA preparations from clinical samples. It allows to detect evenseveral DNA molecules with RCGY site of interest in a presence of excessive amount of methylated DNA. Just 2 simple steps in one tube. More details
Genomic DNA fragmentation with GlaI and subsequent NGS allow to construct a map of R(5mC)GY sites in analyzed genomes of patients and healthy donors. Comparison of these maps reveals R(5mC)GY sites – potential epigenetic markers that can be used for diagnostics. More details
Light fraction of the blood cells is a good source of DNA for our PCR tests. It’s easy to prepare this fraction – one standard table centrifuge, no reagents except saline and minimal time of manipulation with a blood. More details
New type of enzymes – MD DNA endonucleases
Methyl-directed site-specific DNA endonucleases belong to a new type of enzymes discovered by SibEnzyme (Russia).
These enzymes are very similar to restriction enzymes in biochemical properties and cleave DNA completely. A principal difference of new type of enzymes: they cleave only methylated DNA. This property makes MD endonucleases to be a good instrument for epigenetic analysis.
Now 9 different MD DNA-endonucleases are commercially available. Recognition sites of MD enzymes differ in DNA structure and the methylation patterns plus the enzymes may cleave the same recognition site in a different way.
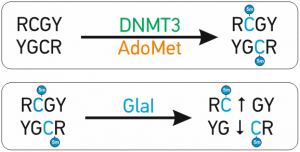
A special advantage of GlaI recognition site R(5mC)GY – this site is the same as a product of DNA methylation with DNMT3, which is responsible for DNA methylation de novo. It makes GlaI to be a very convenient tool for epigenetic diagnostics
Known applications:
- GLAD-PCR assay – for detection of single R(5mC)GY site in point of interest
- EpiSeq – for detection of R(5mC)GY sites in whole genome
Please order MD DNA endonucleases at SibEnzyme site or connect with SibEnzyme managers at info@sibenzyme.com for information how to order at your country.
GLAD-PCR assay – Determination of DNA methylation without bisulfite treatment
GLAD-PCR assay (GlaI hydrolysis and Ligation Adapter Dependent PCR) allows to determine 5′-R(5mC)GY-3′ site in selected position of human and mammalian genomic DNA.
GLAD PCR assay includes three steps and is carried out in one tube [3] (see animation below):
- GlaI hydrolysis of studied DNA
- the universal adapter ligation
- subsequent real-time PCR with Taqman probe.
NO BISULFITE CONVERSION!
A new method of GLAD PCR assay is used to determine a selected 5′-R(5mC)GY-3′ site in a human/mammalian DNA at minimal quantities (5 and more copies) in a presence of excess of unmethylated DNA. It is perfect for epigenetic DNA diagnostics in routine laboratory and clinical practice.
It may be applied to any source of DNA: tissues, biopsies, blood, plasma, serum, etc
GLAD-PCR assay steps
The abnormal de novo DNA methylation is performed by DNMT3A and DNMT3B DNA methyltransferases. These enzymes recognize and methylate site 5’-RCGY-3’ with formation of 5’-R(5mC)GY-3’/3’YG(5mC)R-5’[1]. Recently discovered site-specific methyl-directed DNA-endonuclease GlaI recognizes exactly this DNA sequence 5’-R(5mC)↓GY-3’/3’YG↓(5mC)R-5’ and cleaves it as indicated by arrow [2]












































Innovative aspects and main advantages
In comparison with other methylation detection methods GLAD-PCR has strong advantages:
- Simple – 3 easy steps in one tube and requires only real time PCR-machine
- Quick – only 3-4 hours
- Sensitive – detects from 5 copies of methylated DNA
- Reliable – reaction only in case of methylation – no false positives
- Quantitative – may be used to detect % of methylation
All these factors make GLAD-PCR-Assay to be a very attractive for epigenetic diagnostics
Limitations of traditional methods
The main problem in methylation-specific PCR (MSP), based on bisulfite conversion of DNA, is incomplete conversion which causes false-positive results, or DNA degradation in case of overtreatment.
Methods based on methyl-sensitive restriction enzymes such as pair MspI-HpaII give a positive result if there is no reaction of hydrolysis by HpaII. This is not good for diagnostics as you get positive results in case of absence of reaction which may occur for many reasons, for example in case of non full hydrolysis. And it is not quantative.
Besides all methods require comparison of results of 2 separate reactions.
Primers and TaqMan probes
For GLAD-PCR assay reaction you will need 1 TaqMan probe and 2 primers: genomic and hybrid.
Genomic primer and TaqMan probe located near 5’-R(5mC)↓GY-3’ site of interest are designed as usual [4].
Hybrid primer is a DNA sequence 5’-CCTGCTCTTTCATCGGYNN-3’, wherein 5’-end of
15-nucleotide primer corresponds to universal adapter and tetranucleotide part at the 3’-end (underlined) is complementary to the genomic sequence at GlaI hydrolysis point. This structure implies the existence of 32 variants of hybrid primers corresponding to different possible terminal sequences after GlaI hydrolysis of all possible variants of NNR(5mC)↓GY sequence.
All primers and probes may be ordered at SibEnzyme along with the GLAD-PCR assay kit order. Otherwise, you can order a synthesis of primers and TaqMan probe somewhere else.
GLAD PCR Assay of two R(5mC)GY sites in human genome are used as an example of the method application.
Mixes of TaqMan Probes and primers for analysis of only these two R(5mC)GY sites are included:
- “Control URB1 mix (primers + TaqMan probe)” is provided for analysis of A(5mC)GT site in regulation region of URB1 gene. Position of this site (according to a human genome assembly GRCh38/hg38) is 32334291-32334294 in chromosome 21.
- “Control CEBPD mix (primers + TaqMan probe)” is provided for analysis of G(5mC)GC site in regulation region of CEBPD gene. Position of this site (according to a human genome assembly GRCh38/hg38) is 47738502-47738505 in chromosome 8.
GLAD-PCR assay kit

For researchers convenience we developed a kit for 200 or 1000 GLAD-PCR reactions in 2/10 96-wells PCR plates. All reagents are included except primers and TaqMan probe. The sequences of TaqMan Probe and primers are determined by a customer based on DNA sequence around the RCGY site of interest.
Reaction requires:
- sample DNA
- genomic primer and TaqMan probe designed for RCGY site of interest
- hybrid primer – includes a constant part (complimentary to the universal adapter) and tetranucleotide (complimentary to DNA at the point of GlaI hydrolysis).
Please order GLAD-PCR assay kit at SibEnzyme site or connect with SibEnzyme managers at info@sibenzyme.com for information how to order at your country.
Detailed info on kit in GLAD-PCR assay kit instruction manual
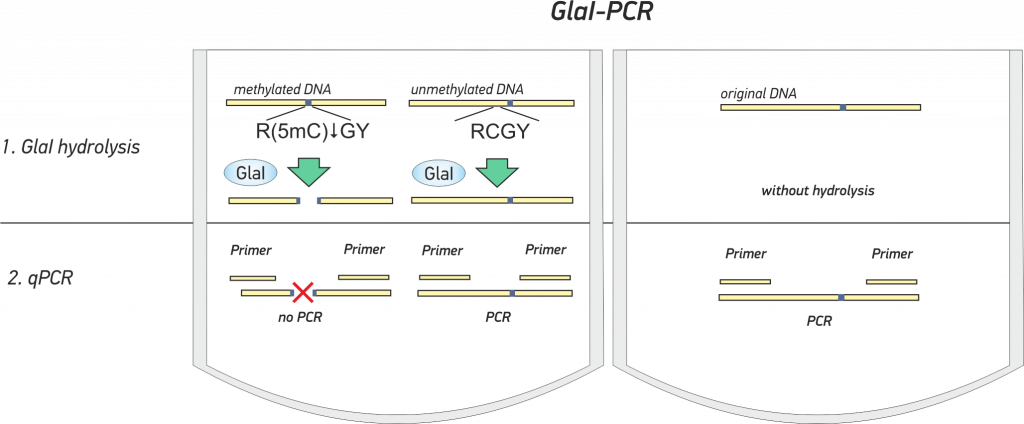
GlaI-qPCR assay – Determination of DNA methylation without bisulfite treatment
GlaI-PCR assay allows to determine 5′-R(5mC)GY-3′ site in selected position of human and mammalian genomic DNA.
GlaI PCR assay includes two steps and is carried out in one tubes [3] (see animation below):
- GlaI hydrolysis of studied DNA
- subsequent real-time PCR with Taqman probe of original and hydrolised DNA
NO BISULFITE CONVERSION!
A new method of GLAD PCR assay is used to determine a selected 5′-R(5mC)GY-3′ site in a human/mammalian DNA at minimal quantities (5 and more copies) in a presence of excess of unmethylated DNA. It is perfect for epigenetic DNA diagnostics in routine laboratory and clinical practice.
It may be applied to any source of DNA: tissues, biopsies, blood, plasma, serum, etc
GlaI-PCR assay steps
The abnormal de novo DNA methylation is performed by DNMT3A and DNMT3B DNA methyltransferases. These enzymes recognize and methylate site 5’-RCGY-3’ with formation of 5’-R(5mC)GY-3’/3’YG(5mC)R-5’[1]. Recently discovered site-specific methyl-directed DNA-endonuclease GlaI recognizes exactly this DNA sequence 5’-R(5mC)↓GY-3’/3’YG↓(5mC)R-5’ and cleaves it as indicated by arrow [2]
Innovative aspects and main advantages
In comparison with other methylation detection methods GlaI-PCR has strong advantages:
- Simple – 2 easy steps in one tubes and requires only real time PCR-machine
- Quick – only 3-4 hours
- Sensitive – detects from 5 copies of unmethylated DNA
- Quantitative – may be used to detect % of methylation
All these factors make GlaI-PCR-Assay to be a very attractive for epigenetic diagnostics
Limitations of traditional methods
The main problem in methylation-specific PCR (MSP), based on bisulfite conversion of DNA, is incomplete conversion which causes false-positive results, or DNA degradation in case of overtreatment.
Primers and TaqMan probes
For GlaI-PCR assay reaction you will need 1 TaqMan probe and 2 primers.

Genomic primers and TaqMan probe located near 5’-RCGY-3’ site of interest are designed as usual [4].
All primers and probes may be ordered at SibEnzyme along with the GlaI-PCR assay kit order. Otherwise, you can order a synthesis of primers and TaqMan probe somewhere else.
GlaI PCR Assay of two R(5mC)GY sites in human genome are used as an example of the method application.
Mixes of TaqMan Probes and primers for analysis of only these two RCGY sites are included
GlaI-PCR assay kit
For researchers convenience we developed a kit for 200 or 1000 GlaI-PCR reactions in 2/10 96-wells PCR plates. All reagents are included except primers and TaqMan probe. The sequences of TaqMan Probe and primers are determined by a customer based on DNA sequence around the RCGY site of interest.
Reaction requires:
- sample DNA
- genomic primers and TaqMan probe designed for RCGY site of interest
Please order GlaI-PCR assay kit at SibEnzyme site or connect with SibEnzyme managers at info@sibenzyme.com for information how to order at your country.
Detailed info on kit in GlaI-PCR assay kit instruction manual
EpiSeq – new method of NGS preparation for DNA methylation profiling
While developing an epigenetic diagnostics the main interest of researchers is positions of de novo DNA methylation sites in the genome – potential methylation biomarkers. De novo DNA methylation is produced by DNMT3A and DNMT3B DNA methyltransferases. These enzymes recognize and methylate 5’-RCGY-3’ site with formation of 5’-R(5mC)GY-3’/3’YG(5mC)R-5’[1]. Thus, there are 4 possible variants of R(5mC)GY site: A(5mC)GC, A(5mC)GT, G(5mC)GC, G(5mC)GT.
A new method of R(5mC)GY sites profiling in the whole genome – EpiSeq developed by SibEnzyme – includes 4 steps:
- DNA fragmentation: GlaI hydrolisys produces fragments with blunt ends
- DNA fragments size select: 150-700 fragments selection
- Illumina paired-end 75-150 bp sequencing using usual reagents
- Fragments mapping to Human genome reference. Fragment ends correlates with R(5mC)GY positions
Detailed method description see in articles [12, 14]
Key facts and advantages
- simple – just replace ultrasound fragmentation by GlaI hydrolysis, fragments ends correspond to DNA methylation points
- 4-6 Gb of data usually is enough for 1 profile; 15-20 samples per 1 HiSeq run
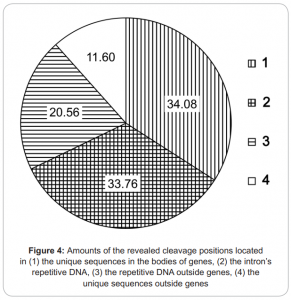
- covers about 3.5 mln of 7 mln possible de novo DNA methylation RCGY sites within genome
Limitations of traditional methods
The most comprehensive data may be obtained with the whole genome bisulfite sequencing (WGBS) method. However, this is rather expensive and laborious technologically; therefore, this method is not used widely.
In practice the less expensive RRBS (reduced representation bisulfite sequencing) method may be applied, which determines sequences of only short, converted MspI genomic fragments, nevertheless covering a big number of CpG islands.
The progress in DNA chip technologies based on bisulfite conversion also should be mentioned. For instance, Infinium MethylationEPIC BeadChip Kit allows to determine the status of methylation for more than 850,000 CG sites in a single experiment. However, the selection of tested bases is too subjective and assay is too expensive.
The common disadvantages of the methods with a bisulfite treatment are a possible incomplete conversion and a high level of DNA degradation. Besides they include several additional stages and are rather sophisticated.
EpiSeq kit is being prepared for orders, GlaI is commercially available. Please order GlaI at SibEnzyme site or connect with SibEnzyme managers at info@sibenzyme.com for information how to order at your country or for more details.
New DNA source from the venous blood
Light fraction of the blood cells is a good source of DNA for our PCR tests. Easy to get this fraction – one standard table centrifuge, no reagents except saline and minimal time of manipulation with a blood. This simplicity allows us to avoid most problems and mistakes of medical stuff which are usual when working with ctDNA or cfDNA. See usage of method at [13]
Conferences, patents and publications
Method presentation at scientific conferences:
- Cancer Epigenetics 2017, Osaka, Japan, October 2017, poster presentation Study of DNA methylation associated with Lung Cancer: GLAD-PCR Assay of R(5mC)GY sites
- Cancer Epigenetics 2017, Osaka, Japan, October 2017, poster presentation Comparative analysis of RCGY sites methylation in the genomes of Raji and U-937 malignant and normal lung fibroblast cell lines
- Global Cancer Summit 2017, Kuala Lumpur, Malaysia, March 2017, oral presentation GLAD-PCR assay of R(5mC)GY sites in aberrantly methylated regulation regions of tumor-suppression genes in lung cancer
- 43th ISOBM annual congress, September, 2016, Chicago, USA, oral presentation GLAD-PCR assay of DNA methylation markers associated with colorectal cancer
- Global Cancer Summit 2015, Bangalore, India, oral presentation “GLAD-PCR assay of R(5mC)GY sites in aberrantly methylated regulation regions of tumor-suppression genes in colorectal cancer“
- Cancer Epigenomics 2015, San-Francisco, USA, poster presentation “Determination of R(5mC)GY sites in regulation regions of tumor-suppression genes by GLAD-PCR assay in colorectal cancer“
- Cell Symposia Cancer Epigenomics, Barselona, Spain, October, 2013, Poster presentation GLAD PCR analysis of aberrant DNA methylation in cancer
Related articles and patents:
- Handa, V., and Jeltsch A. “Profound sequence preference of Dnmt3a and Dnmt3b mammalian DNA methyltransferases shape the human epigenome” // 2005, J. Mol. Biol. 348, 1103-1112
- Tarasova G.V., Nayakshina T.N., Degtyarev S.Kh. Substrate specificity of new methyl-directed DNA endonuclease GlaI // BMC Molecular Biology 2008, 9:7
- Kuznetsov V.V., Abdurashitov M.A., Akishev A.G., Degtyarev S.Kh. Method of determining nucleotide sequence Pu(5mC)GPy at predetermined position of continuous DNA // Patent RU 2525710 C1
- A.G. Akishev, M.A. Abdurashitov, V.L. Sitko, N.A. Netesova, I.F. Radaeva, E.A. Nechaeva, S.Kh. Degtyarev “GLAD-PCR assay of selected R(5mC)GY sites in URB1 and CEPBD genes in human genome” // Res J Pharm Biol Chem Sci, 8(1): pp.465-475, 2017
- A.A. Evdokimov , N.A. Netesova , N.A. Smetannikova , M.A. Abdurashitov, A.G. Akishev , B.S. Malyshev , E.S. Davidovich , V.V. Fedotov , V.V. Kuznetsov , Y.D. Ermolaev , A.B. Karpov , A.E. Sazonov , R.M. Tahauov, S.Kh. Degtyarev GLAD-PCR Assay of DNA Methylation Markers Associated with Colorectal Cancer // Biol Med (Aligarh) , 8:7(2016)
- A.A. Evdokimov, N.A. Netesova, N.A. Smetannikova, M.A. Abdurashitov, A.G. Akishev, E.S. Davidovich, Yu.D. Ermolaev, A.B. Karpov, A.E. Sazonov, R.M. Tahauov, S.Kh. Degtyarev Application of GLAD-PCR assay for determination of the methylation sites in the regulatory regions of tumor-supressors gene ELMO1 and ESR1 in colorectal cancer // Problems in oncology, #1, 2016 p.116-120
- Alexander G. Akishev, Danila A. Gonchar, Murat A. Abdurashitov and Sergey Kh. Degtyarev Epigenetic typing of human cancer cell lines by BlsI- and GlaI-PCR assays // “Ovchinnikov bulletin of biotechnology and physical and chemical biology” V.7, No 3, pp 5-16, 2011
- D. A. Gonchar, A. G. Akishev, S. Kh. Degtyarev BlsI- and GlaI-PCR assays – a new method of DNA methylation study // “Ovchinnikov bulletin of biotechnology and physical and chemical biology” V.6, No 1, pp 5-12, 2010
- V.A. Chernukhin, T.N. Najakshina, M.A. Abdurashitov, J.E. Tomilova, N.V. Mezentzeva, V.S. Dedkov, N.A. Mikhnenkova, D.A. Gonchar, S.Kh. Degtyarev A novel restriction endonuclease GlaI recognizes methylated sequence 5’-G(5mC)^GC-3’ // Biotechnologia (russ.). 2006. N 4. P. 31-35
- A.A. Evdokimov, M.V. Kulak, N.A. Netesova, N.A. Smetannikova, M.A., Abdurashitov, A.G. Akishev, B.S. Malyshev, E.S. Davidovich, V.V. Fedotov, A.B. Karpov, S.Kh. Degtyarev GLAD-PCR Assay of DNA Methylation Markers Associated with Colorectal Cancer // Tumor Biology, Vol.37, supplement 1, Sept. 2016, s6-s7
- Abdurashitov M.A., Tomilov V.N., Gonchar D.A., Dubinin E.V., Degtyarev S.Kh. “Mapping positons of methylated sequences Pu(5mC)GPy along continuous DNA for epigenetic profile creation and identification of aberrant DNA methylation sites” // Patent RU 2586502 C1
- Abdurashitov M.A., Tomilov V.N., Gonchar D.A., Kuznetsov V.V., Degtyarev S.Kh. Mapping of R(5mC)GY Sites in the Genome of Human Malignant Cell Line Raji // Biol Med (Aligarh) Volume 7, Issue 4, BM-135-15 (2015)
- A.G. Akishev, N.A.Netesova, M.A. Abdurashitov, I.V. Vihlyanov, M.K. Nikitin, A.B. Karpov, S.Kh. Degtyarev GlaI-PCR analysis of methylation of ACGC site in Chr11: 65647266 in DNA samples from the blood cells of healthy donors and early stage breast cancers. Epigenet DNA diagnx, vol.1, 2019, DOI: 10.26213/SE.2019.66.12.001
- Abdurashitov M.A., Tomilov V.N., Gonchar D.A., Snezhkina A.V., Krasnov G.S., Kudryavtseva A.V., Degtyarev S.Kh. Comparative Analysis Of RCGY Sites Methylation In Three Human Cell Lines. Epigenetic DNA diagnostics, vol.1, 2019 doi:26213/SE.2019.76.40116 Accessed 11 Nov 2019